Algae
APPLICATIONS
Establishment of a rapid transformation method for wild-type Chlamydomonas strains that does not require cell wall lysis
Chlamydomonas is a eukaryotic unicellular green alga widely used to study many biological findings, including biofuel production.
In this study, we established a method for rapid transformation of wild-type Chlamydomonas strains without removing the cell wall by electroporation using square-wave electrical pulses. By optimizing the electrical conditions, this method is expected to be applicable to the transformation of other industrially useful algae.
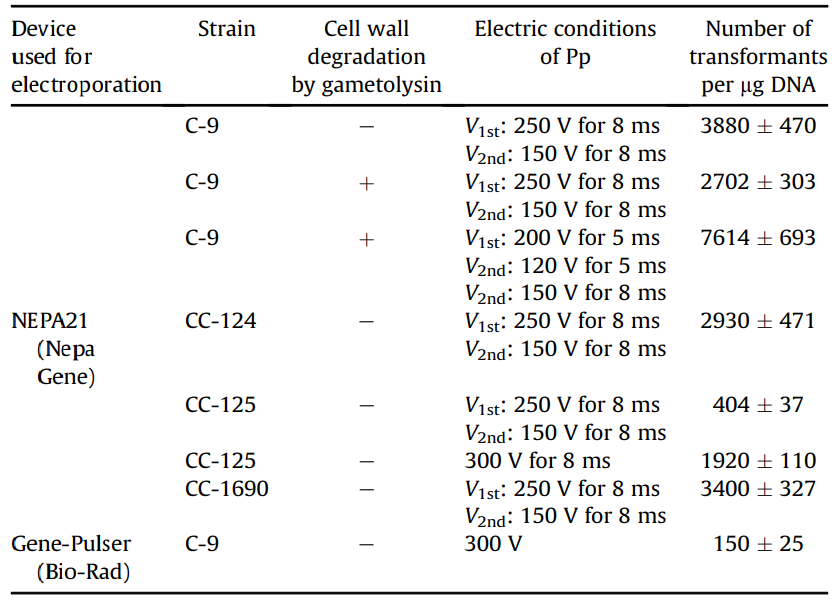
Figure 1: Transformation efficiency of Chlamydomonas using NEPA21
When transforming the Chlamydomonas C-9 strain (with cell wall) using NEPA21, the maximum transformation efficiency was obtained under the following conditions: two Pouring pulses, voltage: 250 V, pulse width: 8 ms, pulse interval: 50 ms, and decay rate: 40%, resulting in 3,880 transformants per 1 µg of DNA when 4 × 106 cells were electroporated. This was approximately 26 times higher than the 150 transformed cells obtained when the existing electroporation method was used to transform the C-9 strains without lysing their cell walls.
In addition, 7,614 transformants per 1 µg of DNA were obtained under optimal conditions for the C-9 strain (protoplasts) in which cell walls were lysed by gammetolysin treatment.
To determine whether this transformation method can be applied not only to the C-9 strain but also to other wild strains, we also examined the transformation efficiency of the commonly used Chlamydomonas wild strains CC-124, CC-125, and CC-1690.
(A): Photograph of a colony of a transformed strain when aphVII cassette (DNA fragment conferring resistance to hygromycin) was introduced into a Chlamydomonas wild-type strain by NEPA21 and spread on TAP agar medium containing 30 µg mL-1 hygromycin B.
(B): Fluorescent image derived from LCIB-GFP of a transformed strain obtained by NEPA21 transfection of pTT1-LciB-GFP. A clear ring-shaped fluorescent signal is observed around the pyrenoid structure. Scale bar: 5 µm
Previous methods required several days to condition the gametolysin prior to transformation. In comparison, with this method, DNA can be introduced directly into wild strains without any prior preparation. By examining the electrical conditions of the pulse, it is also possible to use this method for the transformation of useful microalgae other than Chlamydomonas.
In courtesy of Dr. Takashi Yamano, Dr. Hiro Iguchi and Prof. Hideya Fukuzawa, Laboratory of Applied Molecular Microbiology, Graduate School of Biostudies, Kyoto University
A highly efficient transformation method for the pinnate diatom Phaeodactylum tricornutum using electroporation
A highly efficient transformation method for the pinnate diatom Phaeodactylum tricornutum using electroporation to introduce foreign DNA into cells by multiple sequential pulses has been established. By optimizing the pulse conditions of the NEPA21 electroporator, high transformation efficiency (~4,500 cells per 1×108 cells) was achieved without removing the strong siliceous shell of the diatom.
This is 10 to 100 times higher transformation efficiency than the conventional Pheodactylum transformation method using a particle gun, and the same pPha-T1 vector used in this study. An additional advantage of this method is the short time required to obtain zeocin-resistant colonies. While the particle gun method requires approximately 3 to 4 weeks to produce zeocin-resistant colonies, our method produces colonies within 10 to 15 days after the electric pulse. This may be because the damage to the diatom cells is small and the cell walls remain intact.
Integration and expression of reporter genes into the genome of transformed Pheodactylum cells
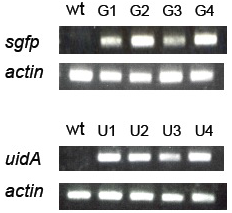

(A): sgfp, uidA, and actin genes in the genomic DNA of transformed Pheodactylum cells were amplified by PCR. Genomic DNA isolated from wild type (wt) and four zeocin-resistant diatom lines (G1-G4 = sgfp transformant, and U1-U4 = uidA transformant) were used as templates for PCR.
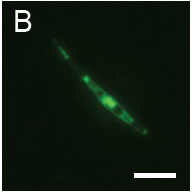
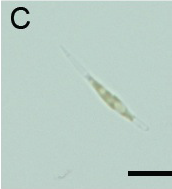
(B): Fluorescence observation and (C): micrograph of Pheodactylum expressing sGFP (S65T) in bright field.
The presence of sGFP(S65T) protein was indicated by bright green fluorescence.
(D): Micrograph of Pheodactylum cells expressing GUS in bright field.
GUS activity is detected by blue staining. Scale bar: 10 µm.
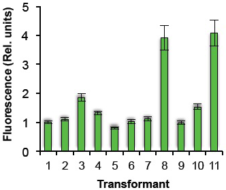
(E): Line-by-line fluorescence intensity comparison of transformed Pheodactylum cells expressing sGFP.
The relative fluorescence intensity of each transformed line was evaluated by Image J. Values: mean ± standard deviation of 5-6 cells
Approximately 90% of the zeocin-resistant colonies expressed the reporter gene, confirming that the expression of the reporter gene remained stable after repeated passages.
In courtesy of Dr. Kentaro Ifuku, Graduate School of Biostudies, Kyoto University and Dr. Yasuhiro Kashino, School of Science, Graduate School of Science, University of Hyogo
Bioscience, Biotechnology, and Biochemistry, Volume 77, Number 4, Pages 874-876, April 2013
PUBLICATIONS
- Phaeodactylum_tricornutum
- Chlamydomonas
Conserved cobalamin acquisition protein 1 is essential for vitamin B12 uptake in both Chlamydomonas and Phaeodactylum
Sayer AP, Llavero-Pasquina M, Geisler K, Holzer A, Bunbury F, Mendoza-Ochoa GI, Lawrence AD, Warren MJ, Mehrshahi P, Smith AG.
Plant Physiol. 2024 Jan 31;194(2):698-714.
- Phaeodactylum_tricornutum
Droplet-based microfluidic screening and sorting of microalgal populations for strain engineering applications
Yu Z, Geisler K, Leontidou T, Young REB, Vonlanthen SE, Purton S, Abell C, Smith AG.
Algal Res. 2021 Jun:56:None.
Electroporation
■ Cell Cultures
- Primary Cell Cultures
- Stem Cells
- Organoids
- Cell Lines
- Cells in Adherence
■ In Vivo Mice/Rats
- Zygotes In Vitro (TAKE method)
- Zygotes In Oviduct (i-GONAD method)
- Embryos In Utero
- Ex Utero Embryos In Vitro
- Brain
- Retina / Cornea / Spinal Cord / Sciatic Nerve
- Lung / Spleen / Liver / Stomach/ Kidney / Intestine
- Pancreas / Islets of langerhans
- Testis / Ovary / Prostate / Gonad / Uterus
- Muscle / Skin / Joint / Cartilage / Tumor / Others
■ In Vivo Other Animals
- Bovine/Porcine/Other Animal Zygotes
- Hamster Zygotes in Oviduct (i-GONAD method)
- Monkey Skin
- Chicken (In Ovo・Others)
- Zebrafish & Other Fishes
- Insects・Others
■ Plant Cells & Algae
- Plant Cells
- Algae
■ Exosomes
- Exosomes
■ Bacteria, Yeast, Fungi
- E. coli/Bacterial Cells
- Yeasts/Fungi
- Bacterial cells/Yeasts/Fungi (NEPA Porator)
Drug Delivery and Transfection
■ Ultrasound Transfection and Drug Delivery (Sonoporation/Fus)
- Brain
- Liver/Skin/Other Applications
- Heart
- Cell Culture
- Lung
- Muscle
Electro Cell Fusion
■ Hybridoma Production
- Monoclonal antibodies, etc
■ Oocyte Activation
- Electrical stimulation before/after Intracytoplasmic sperm injection (ICSI)
■ Somatic cell nuclear transfer (SCNT)・ Oocytes Nuclear Transfer
- Animal cloning
■ Tetraploid Embryos Production
- 2 Cell Embryos (Tetraploid)
■ Other Applications
- Liposome・Protoplast・Yeast, etc.
Fluorescence Quenching / in situ Hybridization Chain Reaction
■ Autofluorescence Quenching
- Mammalian Tissue Sections
- Fish Tissue Sections
- Amphibia Tissue Sections
- Avian Tissue Sections
- Plant Tissue Sections
- Chordate Tissue Sections
■ in situ HCR
- Detection of Target mRNA
Single-Cell/Micro-Particle Transfer
■ マイクロピック&プレースシステム
- Picking and placing micro targets
■ Micro targets
- Animal cells
■ Micro liquid
- Plant cells
Cell Freezing
■ Cell Therapy
- Stem cells, primary cells, and more
■ Animal Husbandry
- Sperm, embryos, tissues, and more